The Drosophila Genomics Resource Center (DGRC) serves the Drosophila research community by: (a) collecting and distributing DNA clones and vectors; (b) collecting and distributing Drosophila cell lines; (c) developing and testing genomics technologies for use in Drosophila and assisting members of the research community in their use.
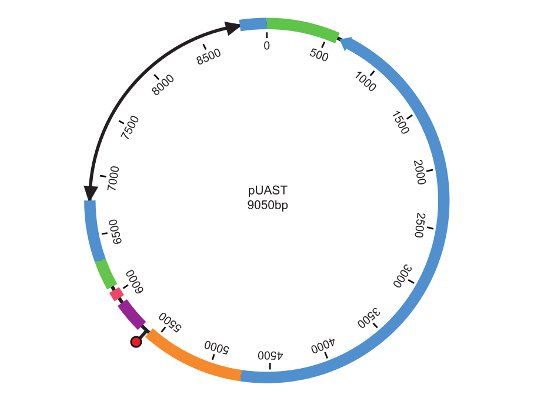
The DGRC has assembled a large (over 1,000,000 and expanding) inventory of DNA clones through the generosity of individual community members, the BDGP, and the CuraGen Corporation. The collection includes transformation vectors, cDNA clones, and the CuraGen yeast two-hybrid collection. Clone collections and common vectors are listed in the catalog.
The DGRC has sought to collect as many cell lines as possible representing D. melanogaster and closely related species. The collection currently includes 242 lines and will continue to grow.
Cell lines, with brief descriptions, are listed in the catalog.
The DGRC is located within the Biology Department of Indiana University, and is supported by a grant from the National Institutes of Health (ORIP and NIGMS). The DGRC was created under the guidance of Dr Peter Cherbas and its work is directed by Dr Andrew Zelhof and an advisory board representing the Drosophila research community.
DGRC User Survey
Let us know at any time how we are doing using our
survey
link or the QR code below. You can also send us feedback directly
at dgrc@iu.edu.